Abstract
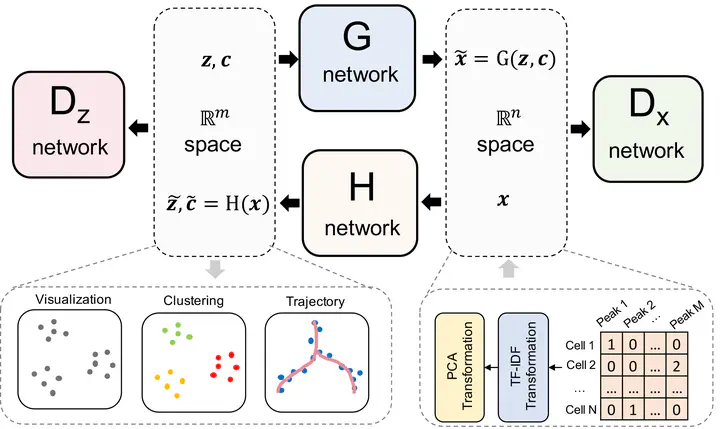
Recent advances in single-cell technologies, including single-cell ATAC-seq (scATAC-seq), have enabled large-scale profiling of the chromatin accessibility landscape at the single-cell level. However, the characteristics of scATAC-seq data, including high sparsity and high dimensionality, have greatly complicated the computational analysis. Here, we propose scDEC, a computational tool for scATAC-seq analysis with deep generative neural networks. scDEC is built on a pair of generative adversarial networks, and is capable of simultaneously learning the latent representation and inferring cell labels. In a series of experiments, scDEC demonstrates superior performance over other tools in scATAC-seq analysis across multiple datasets and experimental settings. In downstream applications, we demonstrate that the generative power of scDEC helps to infer the trajectory and intermediate state of cells during differentiation and the latent features learned by scDEC can potentially reveal both biological cell types and within-cell-type variations. We also show that it is possible to extend scDEC for the integrative analysis of multi-modal single cell data.