Multi-modal Diffusion Model with Dual-Cross-Attention for Multi-Omics Data Generation and Translation
Abstract
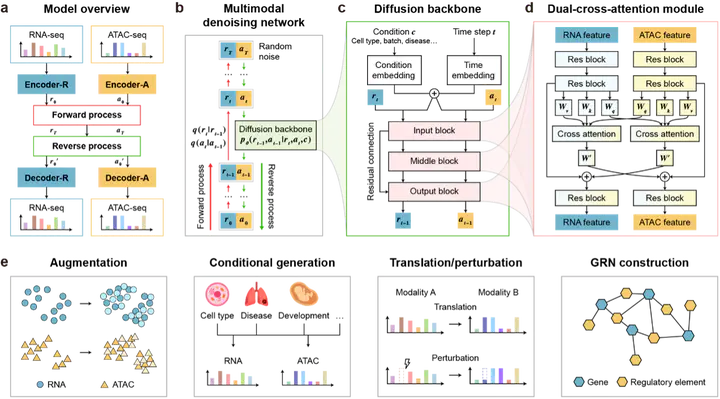
Single-cell multi-omics data have a high potential for deciphering complex cellular mechanisms. But simultaneously measuring multi-omics data from the same cells is still challenging, which calls for computational methods to integrate data of multiple modalities and generate unobserved data. In this paper, we present scDiffusion-X, a latent diffusion model tailored for this task. The model uses autoencoders to map the multi-modalities into low-dimensional latent spaces, coupled with a Dual-Cross-Attention (DCA) module we invented to learn hidden links between modalities. DCA enables the model to unravel interactions among features of multiple modalities and interpretable integration of multi-omics data. We designed a framework with DCA to extract comprehensive cell-type specific heterogeneous gene regulatory network. scDiffusion-X not only excels in generating multi-omics data under various conditions, but also can translate data between modalities with high fidelity, which cannot be achieved with existing multi-omics data simulators. Extensive benchmarking experiments showed that scDiffusion-X has superior performance in scalability, quality of generated data, and model interpretability compared with existing methods. It can serve as a powerful tool for unlocking the potential of single-cell multi-omics data, dissecting regulatory relationships, predicting perturbation responses, and identifying biomarkers and regulatory patterns.
Add the full text or supplementary notes for the publication here using Markdown formatting.